Accelerating Polymer Discovery via Process Intensification
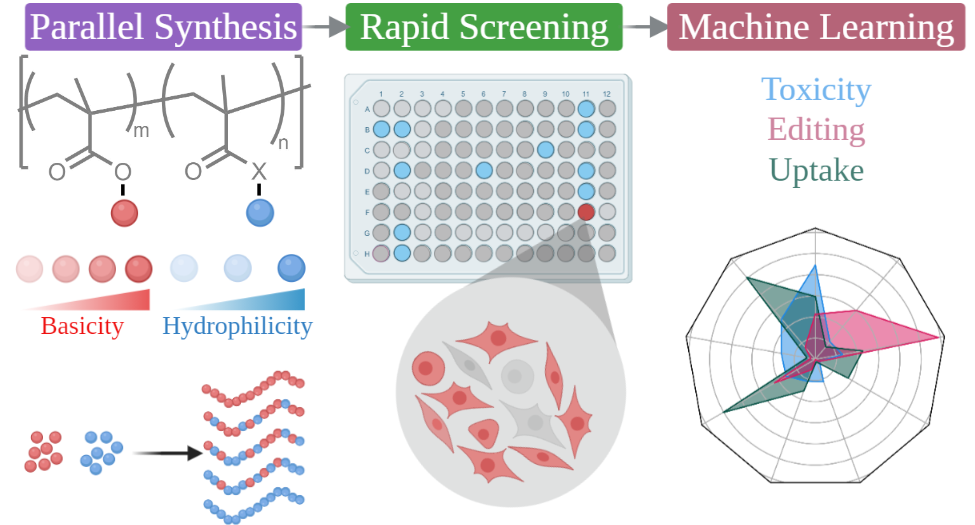
In order to employ data-driven approaches to map relationships between polymer properties and biological responses, we must establish high-throughput experimental (HTE) workflows for biomaterial discovery that opens up unexplored and under-explored chemical domains to investigation. Combining chemical diversity, high discovery rates and the capacity to test multiple mechanistic hypotheses concurrently, HTE approaches will generate large experimental datasets from which structure-activity relationships can be derived via statistical learning. We will exploit synthetic workflows based on continuous flow polymerization and facilitate parallel processing from end to end, right from RDRP, to polymer purification, and finally sample preparation for physicochemical analysis and biological testing.
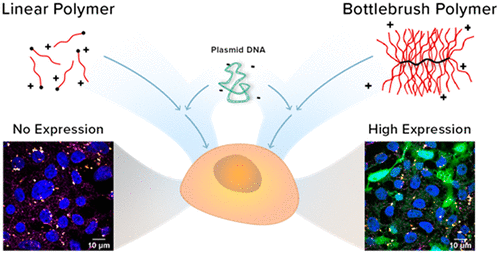
We are also interested in investigating physical pathways for polyelectrolyte complexation (or polyplex formation) by integrating polymer chemistry and process intensification methodologies such as turbulent micromixing to improve polyplex physical properties. By applying process intensification approaches, we seek to decouple physical properties from polymer composition. Some polymer candidates that possess optimal chemical and structural attributes would have been discarded owing to polyplex aggregation. Such polymers may have been excluded from further development without ascertaining whether aggregation resulted from deficiencies in polymer properties or arose instead from the limitations of manual mixing. My lab will redeem some of these interesting polymer candidates by reformulating and re-evaluating aggregation-prone “borderline” polymer candidates via polyplex assembly approaches based on process intensification, thereby bypassing the limitations of manual methods of polyplex assembly.